INTRODUCTION
Cryptosporidium spp. are apicomplexan protozoan parasites that infect a wide variety of vertebrate hosts, including reptiles, birds, fish, amphibians, and mammals, and cause cryptosporidiosis [1]. Cryptosporidiosis is characterized by acute gastrointestinal disturbances, mucoid or hemorrhagic watery diarrhea, fever, lethargy, anorexia, and death in humans and mammals [2]. Cryptosporidium infections have been described in at least 57 reptilian species consisting of 40 species of snakes, 15 species of lizards, and 2 species of tortoises [3]. Unlike in other animals in which infection with Cryptosporidium spp. is usually self-limiting in immunocompetent individuals, cryptosporidiosis in reptiles is frequently chronic and sometimes lethal in snakes [4]. Two species, Cryptosporidium serpentis and Cryptosporidium varanii (syn. C. saurophilum) have been described in snakes and lizards to date [4,5]. C. serpentis is an important parasite in snakes and is usually found in the gastric epithelium [6]. Clinical signs of cryptosporidiosis in snakes have been described as anorexia, lethargy, postprandial regurgitation, midbody swelling, and weight loss [1]. The infection occurs more frequently in adults rather than in young reptiles, unlike in mammals and birds [7]. While C. varanii was originally described in lizards as causing weight loss, abdominal swelling and mortality, it can be found in snakes but has no significant signs [5].
Stressed animals, which have been raised in a limited living space together with various types of species, are more likely to contribute to the spread of the parasite [8]. The above factors have been reported to suppress the immune responses and increase the opportunity for pathogens to cause infections and consequently spread to other animals including humans [8,9]. Cryptosporidium infection in snakes is difficult to identify, especially in those with a subclinical infection [10]. Conventional methods for detection of Cryptosporidium oocysts (including microscopic examination of fecal smears with acid-fast stains) are not capable of identification to the species level. Therefore, molecular techniques have been developed to detect and differentiate Cryptosporidium at the species/genotype and subtype levels [11]. Previously, molecular analysis of Cryptosporidium infection in snakes has identified C. parvum, C. muris, and Cryptosporidium mouse genotype, which probably originated from the ingestion of infected rodents or other prey [4,6,12-14]. Likewise, molecular techniques will help ensure accurate species identification of Cryptosporidium oocysts in snakes.
The pet snake business has become popular in Thailand. However, there is a lack of information regarding Cryptosporidium infection in snakes in Thailand. The present study aimed to identify Cryptosporidium species in captive snake fecal samples using microscopic and molecular examinations. This study will be relevant to disease surveillance and to the improvement of the management of aliments in captive snakes in Thailand.
MATERIALS AND METHODS
Sample collection
In total, 165 fecal samples were collected from asymptomatic snakes of 8 species in 6 genera (Table 1). Of these, 34 snakes were housed in 5 exotic pet shops, and 131 were from 2 snake farms. Fecal samples were stored at 4˚C before analysis.
DNA extraction
DNA was extracted from the supernatant produced using Sheather’s sugar flotation technique with a commercial kit (E.Z.N.A.® Stool DNA Kit, Omega Biotek Inc., Norcross, GA, USA) following the manufacturer’s protocol. DNA was stored at -20˚C before molecular analysis.
Nested PCR amplification and PCR- RFLP analysis
Amplification of the 819-825 bp polymorphic fragment of the SSU rRNA using nested PCR was performed as previously described [17]. Briefly, the PCR conditions were composed of pre-denaturation at 94˚C for 5 min, then 35 cycles of denaturation at 94˚C for 45 sec, annealing at 55˚C for 45 sec, and extension at 72˚C for 1 min, followed by final extension at 72˚C for 10 min. RFLP of the secondary PCR products of C. parvum positive samples was performed using Vsp I (Thermo Fisher Scientific Inc., Rochester, New York, USA) for the genotyping of C. parvum [17]. The reaction mixture contained 0.5 µl of Vsp I (Thermo Fisher Scientific Inc.), 2.2 µl of restriction buffer, and 5 µl of PCR product at 37˚C for 30 min, under conditions recommended by the manufacturer. The digested products were analyzed using 2% agarose gel electrophoresis.
DNA sequencing and phylogenetic analysis
The positive Cryptosporidium samples were submitted for sequencing (1st Base Laboratory, Selangor, Malaysia). The DNA sequences were compared with those in the GenBank database using the basic local alignment search tool (BLAST) algorithm, and the species of Cryptosporidium present in the sample was determined. The nucleotide sequences of the partial SSU rRNA gene of the Cryptosporidium parasites were deposited in the GenBankTM database under the accession nos. KM870564 - KM870603. Multiple alignments were done using the ClustalW program [18,19]. A neighbor-joining tree was constructed from the aligned sequences using the MEGA version 5 software [20].
Statistical analysis
Statistical analysis was performed using a chi-square (χ2) test in the Number Cruncher Statistical System (NCSS) version 2000 to determine the association between the prevalence of Cryptosporidium infection vs host genders and host locations. Values were tested for significance at P≤0.05.
RESULTS
Of the 165 fecal samples from captive snakes, 12 (7.3%) were detected as positive for Cryptosporidium oocysts using the DMSO-modified acid-fast stain, and 40 (24.2%) were positive using nested PCR (Table 1). Seventeen out of 34 samples (50.0%) from pet shops and 23 out of 131 samples (17.6%) from private farms were positive for Cryptosporidium. Among the positive results from nested PCR testing, sequencing analysis identified 24 (60.0%); 9 (22.5%); 3 (7.5%), 2 (5.0%), 1 (2.5%), and 1 (2.5%) as C. parvum, C. serpentis, C. muris, Cryptosporidium mouse genotype, C. andersoni, and C. saurophilum, respectively (Table 1). C. parvum was detected from Boa constrictor constrictor (4), Elaphe guttata (10), Lampropeltis triangulum (2), Python regius (5), and Morelia spilota (3). C. serpentis was detected from Lampropeltis getula (1) and Elaphe guttata (8). C. muris was detected from Python regius. Cryptosporidium mouse genotype was detected from B. constrictor constrictor (1) and P. regius. C. andersoni was detected from M. spilota. C. saurophilum was detected from E. guttata (Table 1).
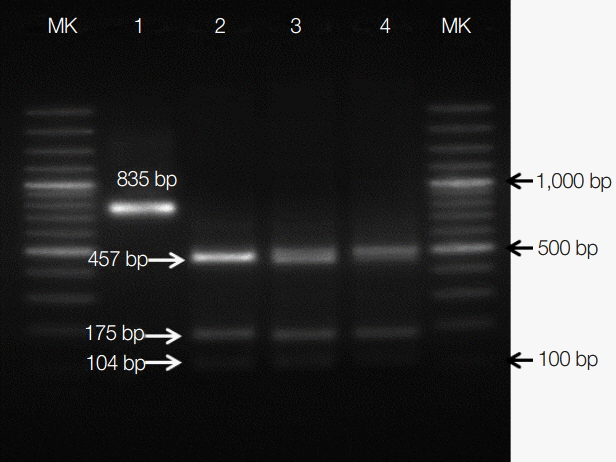
Sequencing analysis indicated that C. parvum, C. serpentis, C. muris, Cryptosporidium mouse genotype, C. andersoni, and C. saurophilum were 99% identical to GenBank accession nos. DQ898158, AF093499, EU553588, EU553589, JX515549, and EU553551, respectively. Differentiation of the C. parvum genotype by PCR-RFLP showed that all 24 C. parvum positive samples were C. parvum mouse genotype (Fig. 1). This genotype is considered as nonpathogenic in snakes [21].
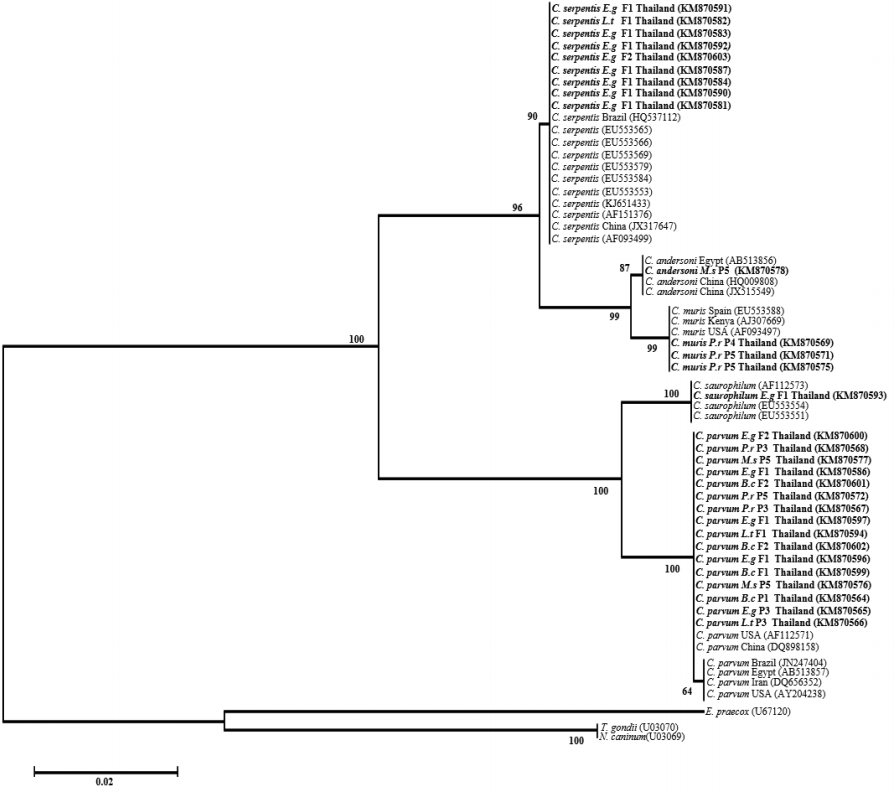
Risk factor analysis showed that there were infection rates of 12.8% and 19.6% of Cryptosporidium spp. in male and female snakes (χ2=1.027, P =0.310), respectively, and the infection rates were 50.0% and 17.6% on pet shops and farms (χ2=17.470, P =0.00008), respectively. These results indicated that location was a statistically significant factor associated with Cryptosporidium spp. infection in snakes (Table 2). The neighbor-joining analysis results showed that 2 distinctive clades of intestinal and stomach Cryptosporidium species from this study were clearly separated (Fig. 2). Intestinal Cryptosporidium species consisted of C. parvum, Cryptosporidium mouse genotype, and C. saurophilum, while stomach Cryptosporidium species consisted of C. serpentis, C. muris, and C. andersoni.
DISCUSSION
Cryptosporidiosis is a well-known cause of chronic hypertrophic gastritis, and is possibly lethal in captive snakes [5,22]. The present study was the first identification of Cryptosporidium infection in pet snakes in Thailand. We found 7.3% of the Cryptosporidium oocysts using the acid-fast stain technique and 24.2% of the Cryptosporidium DNA using a molecular method. The acid-fast staining technique is less sensitive for the detection of Cryptosporidium infection. However, it is the easiest and most effective method for veterinary clinical diagnosis [23]. Moreover, PCR is a sensitive and specific detection technique, which resulted in a much higher proportion of Cryptosporidium positive samples in comparison to acid-fast staining [24]. Additionally, PCR is an important method for the identification of Cryptosporidium species or genotypes in samples with low numbers of oocysts [25,26]. Thus, PCR is still hindered by its high cost and time-consuming DNA extraction, PCR amplification, and gel electrophoresis [27,28]. A combination of several diagnostic techniques for the detection of Cryptosporidium oocysts is still needed [29].
In the present study, sequencing analysis of the 18S rRNA gene revealed the presence of C. serpentis in the corn snake (E. guttata) and king snake (L. getula). These findings were similar to previous studies, which have found that C. serpentis is most common in snakes [4,6,9,12,14,30]. Additionally, we found C. saurophilum in the corn snake (E. guttata), which is similar to previous reports [4-6,13]. C. saurophilum was originally described as an intestinal parasite mainly in lizards [31]. The presence of C. saurophilum in reptiles other than lizards might have resulted from the fact that they were housed together [4-5]. Moreover, none of the infected animals showed any clinical signs of the disease in the present study, which was in agreement with previous reports [6,9]. However, the subclinical stage can last for years in these animals [32].
We have confirmed the presence of C. parvum, C. muris, and C. andersoni in captive snakes. C. parvum and C. muris were considered as Cryptosporidium from mammals. Therefore, they were not pathogenic in snakes [33,34]. On the other hand, we have also confirmed C. andersoni in captive snakes. Interestingly, C. andersoni was commonly found in infected cattle abomasum. Possibly, the finding of other Cryptospodium genotypes in snakes might be due to infection from infected prey animals [35]. This is the first report of C. andersoni in captive snakes.
The study of farm management found that the sanitary conditions on the farm and inadequate management influenced the rate of infection of Cryptospodium in dairy cows [36]. Farms with poor management and bad sanitary conditions present a high risk of Cryptospodium and other gastrointestinal protozoan infections. In this study, risk factor analysis indicated that location was significantly associated with Cryptospodium spp. infection. The risk of infection was higher in pet shops than in farms even though the former did not use for breeding, but only for selling snakes. However, our observation during sample collection found that some pet shops have many species of animals, and keep them at high densities in small cages. They also used contaminated equipment. The result is that pet shops tend to have a higher percentage of Cryptospodium spp. infection than private farms.
Phylogenetic analysis of SSU rRNA gene fragments within the genus Cryptospodium has proven to be a useful tool for both the systematic analysis of the presently recognized species and the possibility of definitive identification of new species or genotypes within this genus [4]. In this study, we identified 6 different Cryptospodium species or genotypes in reptiles. These results confirmed that C. serpentis and C. parvum are the main species found in snakes.
A high proportion of Cryptospodium species from mammals (75.0%) was detected in this study consisting of C. parvum, C. muris, Cryptospodium mouse genotype, and C. andersoni. However, these Cryptospodium are non-pathogenic in snakes but potentially zoonotic. Cryptosporidiosis causes mucoid or hemorrhagic diarrhea, fever, lethargy, anorexia, and death especially in immunocompromised patients [11,37-39]. Additionally, C. muris is probably the zoonotic Cryptospodium, which was reported in HIV patients in Perú, Thailand, Indonesia, France, and Kenya [40]. Recently, C. andersoni has been found in 21 diarrhea patients out of 232 outpatients in China, whereas Cryptospodium hominis (the human genotype) was found only in 2 patients [41]. In 2015, C. andersoni was first reported in a captured lesser panda in China [42]. Therefore, the present identification of C. andersoni might have a public health impact. Additionally, Cryptospodium has an environmentally resistant oocyst, which is a public health risk factor for handlers and owners, especially when they are children, elderly people, or immunocompromised patients. Consequently, feeding snakes with Cryptospodium infected mice or any other prey could possibly transmit the pathogen to humans via feces, water, and contaminated equipment. Although a substantial study of Cryptospodium transmission from reptiles to humans has not been reported yet, disinfection processes have been recommended as the best option for reducing the transmission risk by the application of 4-cholr-M-cresol, 5% ammonium solution, or hydrogen peroxide-based disinfectants [14,21,32]. The presence of Cryptospodium infection in snakes should not be ignored in snake collections because infection can be transmitted from animal to animal by the fecal-oral-route when animals are housed together. The lack of effective treatment for cryptosporidiosis almost always results in euthanasia of the infected snakes, which often leads to the loss of valuable animals in a collection [43]. Hyperimmune bovine colostrum (HBC) is recommended to combat clinical and subclinical C. serpentis infections in captive snakes [44].
In conclusion, applying a molecular method for the detection of the pathogen will probably prove the presence of the Cryptospodium species or genotype, because it is difficult to identify oocysts of pathogenic C. serpentis or C. saurophilum from those of non-pathogenic Cryptospodium species [45]. The study has shown that molecular techniques can separate different species and genotypes of Cryptospodium. The sequencing of the PCR products revealed that C. parvum, C. serpentis, C. muris, C. mouse genotype, C. andersoni, and C. saurophilum are different from each other. In addition, the PCR-RFLP technique can differentiate various C. parvum genotypes (e.g., C. parvum human, C. parvum mouse, and C. parvum bovine genotypes) [45]. This study found a high percentage of non-pathogenic Cryptospodium in snakes in Thailand. However, some species were zoonotic Cryptospodium, which might have been ingested from prey and passed through intestinal tract of the snake.
Cryptospodium oocysts from food items can cause the misidentification of cryptosporidiosis in snakes. In addition, paying more attention to cryptosporidiosis in snakes is required due to public health concerns. Moreover, the sanitary conditions associated with snake feeding should be improved along with avoiding the purchase of infected feeds. Thus, practicing good sanitation and hygiene, including at snake prey suppliers, should be considered, and these places tested for specific pathogens and to prevent the killing of snakes infected with non-pathogenic Cryptospodium.