Identification of parasite DNA in common bile duct stones by PCR and DNA sequencing
Article information
Abstract
We attempted to identify parasite DNA in the biliary stones of humans via PCR and DNA sequencing. Genomic DNA was isolated from each of 15 common bile duct (CBD) stones and 5 gallbladder (GB) stones. The patients who had the CBD stones suffered from cholangitis, and the patients with GB stones showed acute cholecystitis, respectively. The 28S and 18S rDNA genes were amplified successfully from 3 and/or 1 common bile duct stone samples, and then cloned and sequenced. The 28S and 18S rDNA sequences were highly conserved among isolates. Identity of the obtained 28S D1 rDNA with that of Clonorchis sinensis was higher than 97.6%, and identity of the 18S rDNA with that of other Ascarididae was 97.9%. Almost no intra-specific variations were detected in the 28S and 18S rDNA with the exception of a few nucleotide variations, i.e., substitution and deletion. These findings suggest that C. sinensis and Ascaris lumbricoides may be related with the biliary stone formation and development.
The biliary stone disease is a common disorder, which is usually associated with stones in the common bile duct and gallbladder, and can induce significant complications. Stone formations generally occur in the presence of the following factors: abnormalities of the bile constituents, bile stasis, and the presence of nidus (Chong, 2005). Among these factors, biliary parasites are known to be relevant to many aspects of biliary stone diseases. Biliary parasites induce necrosis, inflammation, fibrosis, strictures, and cholangiectasis of the bile ducts via several mechanisms: (1) as a direct result of the irritating chemical composition of the parasite, parasite secretions, and eggs, (2) physical obstruction of the bile ducts, (3) induction of the formation of biliary stones, and (4) introduction of bacteria into the biliary system during migration of parasites from the duodenum. Therefore, bacterial cholangitis plays an important and frequently dominant role in the pathogenesis and clinical course of biliary diseases as the result of these parasitic infestations (Carpenter, 1998). Common biliary parasites include the nematode Ascaris lumbricoides (Choi et al., 1993), and the trematodes Opisthorchis viverrini (Sripa et al., 2004) and Opisthorchis felineus (Sripa et al., 2004), Clonorchis sinensis (Lee et al., 1992), and Fasciola hepatica (Gulsen et al., 2006), and the metacestodes of Echinococcus granulosus and Echinococcus multilocularis (Shemesh et al., 1986). Among these parasites, A. lumbricoides and C. sinensis are the most prevalent and clinically significant parasitic infections detected in the biliary system (Leung and Yu, 1997). However, there have been no reports thus far on the amplification of parasite DNA from biliary stones. We hypothesized that parasite DNA might exist in the pigmented bile duct stones. The objective of the present study was to identify parasite DNA in human biliary stones via the polymerase chain reaction (PCR) and DNA sequencing.
A total of 20 biliary stones, consisting of 15 common bile duct stones and 5 gallbladder stones, collected from 20 patients (one stone per each patient) were examined in the present study (Table 1). The patients were those who were admitted to the Department of Internal Medicine, Kangdong Sacred Heart Hospital, Hallym University, between September and December 2005. All stones were collected during ERCP (endoscopic retrograde cholangiopancreatography) or biliary tract surgery in patients with common bile duct stones or gallbladder stones. The stones were then stored at -70℃ until assayed. We also collected patient histories, especially with regard to the consumption of raw freshwater fish as an intermediate host for C. sinensis.
For mechanical breaking, all the stones were frozen in liquid nitrogen gas for 10 min and thoroughly dried and minced in jars for 5-10 min. Genomic DNA was isolated by a Wizard genomic DNA purification kit (Promega, Madison, Wisconsin, USA) via a modified animal tissue protocol, in accordance with the manufacturer's instructions.
PCR was conducted using a mixed solution of the extracted DNA as 10-30 ng of template DNA, 1.5 µl of dNTP mixture (each 2.5 mM), 0.5 µl of primer (each 10 pmole/µl), 2.0 µl of 10 x Ex Taq buffer, and 0.5 µl of Ex Taq enzyme (5 U/µl, TaKaRa Ex Taq Kit, Takara Shuzo Co., Tokyo, Japan) in a GeneAmp PCR System 2400 (Applied Biosystems, Foster City, California, USA). The PCR reaction cycle for 28S rDNA consisted of 50 cycles of denaturation at 94℃ for 1 min, annealing at 52℃ for 45 sec, and extension at 72℃ for 45 sec, followed by a 10 min final extension. The 28S rDNA PCR primers (JB10 and JB9) were used as reported by Qu et al. (1988). The PCR primers (forward primer; Asc10, reverse primer; Asc11) for the 18S rDNA gene were designed for the amplification of a fragment based on the alignment of published Ascaridoidea sequences by Loreille et al. (2001). The 18S rDNA PCR was conducted using the above described method, but with different primers. The PCR condition utilized was identical to that described above, except that annealing was conducted for 45 sec at 55℃. The PCR product was then run on 2% agarose gels and stained with ethidium bromide.
For cloning and sequencing of 28S D1 and 18S rDNA, the PCR products amplified with the primer sets were purified via gel extraction (Qiagen, Valencia, California, USA) and were subcloned into pGEM-T-easy vector (Promega) plus ligase and JM109 competent cells (Promega), in accordance with the protocols of the supplier. The recombinant plasmid was screened with isopropyl-β-thiogalactoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal). The cloned fragments in the recombinant plasmids were digested with the EcoRI enzyme and purified from agarose gel using a QIAprep spin plasmid kit (Qiagen). DNA sequencing was conducted via the dideoxy chain termination method, using an automated DNA sequencer (Model 373A, Applied Biosystems, Foster City, California, USA) with T7 primers by the COSMO company (Seoul, Korea). At least 3 clones were sequenced per sample with additional clones sequenced as required to resolve ambiguous sites.
Sequence analysis was conducted using the nucleotide BLAST program with the NCBI database (National Center for Biotechnology Information, NIH, Bethesda, Maryland, USA) for similarity and nucleotide length. All sequences obtained were then automatically aligned using the CLUSTAL W program (version 1.82, CLUSTAL W WWW Service at the European Bioinformatics Institute, http://www.ebi.ac.uk/clustalw) for multiple sequence alignments. The resultant aligned output was adjusted manually in order to improve the homology statements. The confirmed sequences for each species were then deposited in the EMBL/GenBank Data Libraries of the NCBI (GenBank No. EF654661 for 28S D1 rDNA gene of C. sinensis, and No. EF654660 for 18S rDNA gene of A. lumbricoides).
To assess the presence of C. sinensis or other helminth eggs from the experimental samples from the 3 and/or 1 positive patient, stool examinations and intradermal tests were conducted. The stool specimens were examined via a direct smear technique. For the skin test, the C. sinensis and Paragonimus westermani antigen detection kit (Green Cross, Yongin, Gyeonggi-do, Republic of Korea) was used. Each antigen (0.01-0.02 ml) was injected intradermally into the volar surface of the forearm. Wheals of an average diameter of 60 mm2 or more emerging in 15 min were considered as positive reactions (Joo et al., 1997).
The demographic characteristics of the patients with common bile duct stones and gallbladder stones are summarized in Table 1. The 15 common bile duct stone patients suffered from cholangitis without evidence of cholangiocarcinoma. The 5 patients with gallbladder stones evidenced acute cholecystitis. Using the 2 primer pairs for the 28S rDNA and 18S rDNA genes, the PCR product was successfully obtained from 3 and/or 1 common bile duct stone extract from the total 15 common bile duct stones (Fig. 1). The size of the 28S D1 rDNA gene product showed an approximately 350 bp, and the size of 18S rDNA gene product yields an approximately 250 bp (Fig. 1). For C. sinensis, the 28S D1 rDNA genes were amplified from 3 positive patient stones (lane 1: a 90-year-old female, lane 7: a 69-year-old male, lane 8: a 51-year-old male) among the 15 common bile duct stones (Fig. 1A). For A. lumbricoides, the 18S rDNA gene was amplified from 1 positive patient stone (lane 7: a 69-year-old male) among 15 common bile duct stones (Fig. 1B). The size of each PCR product was the same within the same gene (Fig. 1). Both genes were amplified from the 1 common bile duct patient stone (lane 7) (Fig. 1). However, no amplification was revealed from the 5 gallbladder stone extracts in both genes (data not shown).
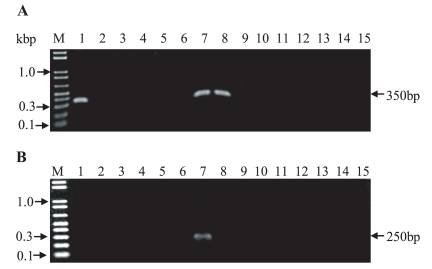
PCR amplification of 28S D1 (A) and 18S rDNA (B) gene fragments from DNA extracted from 15 human common bile stones. The samples were separated via electrophoresis on 2% agarose gel. Lane M: size marker (0.4 µg, 1 kbp plus DNA ladder, Invitrogen). In panel A; Lane 1, 7 and 8: 28S D1 rDNA gene for C. sinensis positive patients (1: a 90-year-old female, 7: a 69-year-old male, 8: a 51-year-old male), In panel B; Lane 7: 18S rDNA gene for A. lumbricoides positive patients (7: a 69-year-old male).
The PCR products were cloned and sequenced from 3 and/or 1 common bile duct stones. All of the sequences acquired from the various clones were well preserved. The nucleotide size of the 28S D1 rDNA gene yielded a 297 bp sequence (Fig. 2), and the size of 18S rDNA gene product yielded an approximately 147 bp sequence (Fig. 3). Each of the nucleotide sequences was the same within the same gene (data not shown). The 28S D1 rDNA sequences were compared with the sequences of C. sinensis (AF188121; Korea isolate, AF217096; China isolate) (Lee and Huh, 2004) and O. viverrini (AF408149) in GenBank (Fig. 2). Our stone sequence and the Korean C. sinensis isolate sequence revealed perfect identity, whereas 7 nucleotide deletions (Fig. 2) (identity 290/297 bp; 97.6%) were detected with the China C. sinensis isolate, and 2 substitutions (identity 295/297 bp; 99.3%) were detected with the O. viverrini sequence (Fig. 2). The 18S rDNA sequences were compared with the sequences from the family Ascarididae, including A. lumbricoides, Ascaris suum, and Toxascaris leonina in GenBank (Fig. 3). A high level of identity was observed between our stone sequence and other Ascarididae sequences; only 2 substitutions (identity 145/147 bp; 98.6%) were detected in comparison with A. suum (AF036587) isolates and the T. leonina sequence (V94383), and 3 substitutions (identity 144/147 bp; 97.9%) were detected in comparison with the A. lumbricoides sequence (Fig. 3). Of the 15 common bile duct stone patients, the positive patients were examined their stools for parasitic eggs by the formalin-ether sedimentation method. However, no parasite eggs (for example, C. sinensis and A. lumbricoides) were detected. The results of the skin test were negative. No blood eosinophilia was detected in the 3 and/or 1 positive patients. The subjects had no history of consuming raw freshwater fish and no past history of Clonorchis infection among the 20 biliary stone patients.

Nucleotide sequences of a region of the 28S D1 rDNA gene of Clonorchis sinensis from the common bile duct stone extracts compared to modern sequences among C. sinensis and O. viverrini. A dot (.) denotes a same identical nucleotide position. A non-identical nucleotide denotes nucleotide addition in same position. Each of primer sequences on both sides is indicated by an underline. Alignment gaps are indicated by a hyphen. Sequences for each species have been deposited in the GenBank databases (GenBank accession No.: C. sinensis; EF654661 (stone), AF188121 (K; Korea isolate), AF217096 (C; China isolate), O. viverrini (AF408149).

Nucleotide sequences of a region of the 18S D1 rDNA gene of Ascaris lumbricoides from the common bile duct stone extracts as compared to modern sequences among the family Ascarididae. A dot (.) denotes an identical nucleotide position. A non-identical nucleotide denotes nucleotide addition at the same position. Each of the primer sequences are indicated by an underline. The sequence is from GenBank (EF654660; A. lumbricoides from stone, U94366; A. lumbricoides from human, AF036587; A. suum from pig, and U94383; Toxascaris leonina from dog).
Numerous investigators have suggested that parasites play a role in human biliary stones (Chai et al., 1991; Choi et al., 1993), but the molecular biological evidence in this regard remains unclear. Thus far, there was not detected DNA amplification from biliary stones. In the present study, we successfully isolated parasite DNA from the common bile duct stone. C. sinensis and A. lumbricoides DNA were found in 3 and/or 1 of 15 common bile duct stones. The 28S and 18S rDNA genes were selected due to their high copy numbers, and also because they are well-known markers for elucidation of evolutionary relationships (Lee and Huh, 2004). It was suggested that no mixed infections of 2 different parasite species existed in the samples. However, 1 patient evidenced co-infection with C. sinensis and A. lumbricoides based on the PCR and sequencing results. Although stool examinations and skin tests are helpful for the diagnosis of clonorchiasis and/or ascariasis (Lee et al., 1992), no parasite eggs were detected in the stool and the skin test was negative. It can be speculated that the exogenous parasite DNA may have been introduced into the hepatic stone a long time ago. Although it remains uncertain that both parasite DNA originated solely from dead Clonorchis and Ascaris specimens, our results showed that common bile duct stones clearly were, at least in part, the result of parasites. This PCR-based molecular approach may eventually facilitate a greater understanding of stone pathogenesis.
