AbstractTo confirm that Bartonella and Wolbachia were carried by sheep keds (Melophagus ovinus) in southern Xinjiang of China, 17 M. ovinus samples, which were collected in Aksu Prefecture, Xinjiang, were randomly selected. In this study, the Bartonella gltA and Wolbachia 16S rRNA gene were amplified through conventional PCR and the sequence of those amplified products, were analyzed. The results demonstrated that Bartonella was carried by all of the 17 sheep keds and Wolbachia was carried by 15 out of them. Bartonella was identified as B. melophagi. Three strains of Wolbachia were supergroup F and 1 strain has not been confirmed yet. It is the first report about Wolbachia supergroup F was found in sheep keds and provided the molecular evidence that B. melophagi and Wolbachia supergroup F were carried by sheep keds in Aksu Prefecture of southern Xinjiang, China. The 2 pathogens were found in sheep keds around Taklimakan Desert for the first time.
INTRODUCTIONSheep keds, a kind of ectoparasites from Hippoboscidae (Diptera: Hippoboscoidea) that subsist solely on blood [1,2], parasitize on sheep. However, they have also been reported parasitize on other domestic and wild animals, including goats [3], rabbits [1], dogs [4], Tibetan antelope [2,5], European bisons [6], red foxes [7] as well as humans [4].
Sheep keds have been widely distributed geographically. It was originally found in most parts of Europe, Northwest Africa, Mongolia and northern India, and later found in Kenya, South Africa, Japan, Australia, New Zealand and most parts of North America [2]. However, sheep keds parasitizing on sheep and Tibetan antelopes were only reported in few areas in China such as Xinjiang [2,8], Qinghai [2,5], Liaoning [9], and Gansu [10,11]. In addition, adult and pupae of sheep keds were found on the imported sheep, sheep skin and wool in quarantine ports of Shandong [12], Beijing [13], and Zhejiang [14].
Sheep keds could cause damage to skin, inflammation, anemia, slow weight gain, poor wool quality, and low yield for sheep [1,2,10,15]. Moreover, it is a media for insect-borne pathogens such as Trypanosoma melophagium [16], Anaplasma ovis [17], blue-tongue virus [18], Bartonella schoenbuchensis, Bartonella chomelii [19], B. melophagi [15,20], and Bartonella spp. In China, Borrelia garinii and Borrelia valaisiana-like groups [8] were detected in sheep keds on Tibetan; Rickettsia raoultii and Rickettsia slovaca were detected in sheep keds on Xinjiang [2]; Bartonella, Arsenophonus, Wolbachia [10,11], Enterobacter, Shewanella, Acinetobacter, Bacillus, Halomonas, and Staphylococcus [10] were detected in sheep keds on Gansu. Sheep keds have caused significant economic losses to the world’s animal husbandry.
Bartonella initially belongs to Rickettsiales, Bartonellaceae. According to later genetic analysis, it then belongs to Rhizobiales, Bartonellaceae, Bartonella. It is a widely distributed facultative intracellular parasite and transmitted by hematophagous arthropods among vertebrates [16]. At least 30 Bartonella that cause disease in human body have been reported so far [15]. Nowadays, B. bovis, B. melophagi, B. capreoli, B. chomelii, B. dromedarii, and B. schoenbuchensis have been identified in cloven-hoofed animals [20]. Wolbachia spp. belongs to Rickettsiales, Anaplasmataceae, Wolbachia. It is the most widely distributed intracellular symbiotic bacteria in the world. About 40% of arthropods and over 65% of insect species naturally carry Wolbachia [21,22]. At present, phylogenetic analysis based on multiple genes was used to divide Wolbachia into 16 supergroups [23]. It has been given wide attention because it can participate in the reproductive regulation of arthropod hosts in various ways.
Bartonella and Wolbachia are distributed globally. They are rarely studied in China and widely distributed in Xinjiang. It is necessary to detect the pathogens of sheep keds such as Bartonella and Wolbachia. It will make an important contribution to the health of humans and animals.
MATERIALS AND METHODSSheep keds samplesIn July 2016, about 300 sheep keds specimens were collected from sheep of local fair in Yaha Town of Kuqa in Aksu, Xinjiang (1,029 m above sea level; 41°44′N, 83°14′E). Seven experimental specimens were selected from them and preserved in 70% ethanol.
In June 2017, more than 200 sheep keds specimens were collected from each of the 5 sheep from the local fair in Yaha Town of Kuqa in Aksu, Xinjiang. These sheep keds specimens were placed in sampling vials with sufficient air and transported immediately to the laboratory for cryopreservation. Two sheep keds specimens were randomly selected from each sheep as experimental specimens.
In this study, 17 samples were processed individually.
DNA extraction, PCR and sequence analysisSheep keds preserved in 70% ethanol ware washed by using 50% ethanol, 30% ethanol, 10% ethanol. And they were followed by washing with distilled water. The cryopreserved sheep keds were washed twice with distilled water. After the elution, they were dried by sterile filter paper and cut into pieces, and then placed into 2 ml sterile tubes. The kit was operated according to instructions of TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver. 5.0 (TaKaRa, Dalian, China). And then the sheep keds genomic DNA was extracted and eluted with 50 μl Elution Buffer twice. Finally, the genomic DNA was collected and then stored at −20°C.
The Bartonella gltA gene and the Anaplasma 16S rRNA gene were amplified according to the instructions of the kit of Premix Taq™ (TaKaRa Taq™ Version 2.0) (TaKaRa). Bartonella gltA gene amplification primers were 5′-GGG GAC CAG CTC ATG GTG G-3′and 5′-AAT GCA AAA AGA ACA GTA AAC A-3′ [24,25]; the expected amplification product size was 379 bp. The genus-specific set of primers for 16S rRNA gene were used for Wolbachia DNA amplification, and the primers sequence were 5′-GGT ACC YAC AGA AGA AGT CC-3′ and 5′-TAG CAC TCA TCG TTT ACA GC-3′ [26]; the expected amplification product size was 345 bp. The PCR system was 50 μl and the annealing temperature was 55°C and 54°C, respectively.
After the amplification products were identified by 1% of gel electrophoresis, the positive amplification products were sequenced using the ABI PRISM™ 3730 XL DNA Analyzer. Sequencing results were performed using the BLAST online platform (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), Primer Premier 5.0, DNAMAN, DNAStar, and Molecular Biology Software (MEGA 5.0) to conduct the sequence analysis and phylogenetic tree construction. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model.
RESULTSDetection of B. melophagiPCR was conducted on 17 sheep keds samples from Kuqa County, Aksu Prefecture, Xinjiang using the Bartonella gltA gene amplification primers, and all samples were positive (100%, 17/17). The size of the amplified products was identical as expected. The amplified products of gltA gene from randomly selected 5 sheep keds were sequenced. The sequences results were analyzed and compared by the BLAST. The results were over 99% in similarity when compared with the Bartonella gltA gene sequence in the database. Moreover, the top 5 gene sequence hosts in similarity were all ectoparasites of Hippoboscidae (Diptera: Hippoboscoidea) (Table 1). A total of 42 nucleotide sequences based on Bartonella gltA gene (312 bp) and Rickettsia gltA gene (330 bp) of outgroup sequence showed that 5 segments of the Bartonella gltA gene sequences collected in this study was clustered together with other B. melophagi gltA gene sequences from other sheep keds hosts (Fig. 1). In this study, 5 Bartonella gltA gene fragments from Aksu Prefecture, Xinjiang were named as B. melophagi XJNJ-1, XJNJ-2, XJNJ-3, XJNJ-4, and XJNJ-5 strains. The accession numbers were MG701233, MG701234, MG701235, MG701236, and MG701237, respectively, when submitting to GenBank.
Sequence analysis of Wolbachia of sheep kedsPCR was conducted on 17 sheep keds samples from Kuqa County, Aksu Prefecture, Xinjiang using the Anaplasma 16S rRNA gene amplification primers. The results showed that 15 samples were positive with the positive rate at 88.23% (15/17) and the size of the amplification products was consistent as the expected size. The 16S rRNA gene amplification products from 5 randomly selected sheep keds were sequenced and then 4 sequence results obtained were compared and analysised by the BLAST. Three segments of them have the highest similarity at 99% after comparing with Wolbachia 16S rRNA gene sequences in the database. Another segment has the similarity at 96%. The top 5 gene sequence in similarity all contain 2 segments of the gene sequences from the host sequence of sheep keds (Table 1). A total of 40 nucleotide sequences of supergroups A-N based on Wolbachia 16S rRNA gene (247 bp) and Rickettsia 16S rRNA (250 bp) of outgroup sequence showed that 4 segments of Wolbachia 16S rRNA gene sequence collected in this study was clustered together with the Wolbachia sequence of 2 sheep keds hosts and Wolbachia sequence of other supergroup F (Fig. 2). The 3 segments designated as supergroup F with 1 segment need further analysis with other genes. In this study, 4 Wolbachia 16S rRNA gene fragments from sheep keds collected from Aksu Prefecture of Xinjiang were named as Wolbachia XJNJ-1, XJNJ-2, XJNJ-3, and XJNJ-4 strains; accession numbers were MG701229, MG701230, MG701231, and MG701232, respectively, when submitting to GenBank.
DISCUSSIONFrom routine PCR, sequencing, and sequence analysis on sheep keds in Aksu Prefecture of southern Xinjiang, China, the results showed that the Bartonella was carried by all the 17 sheep keds and the Wolbachia was carried by 15 out of them. According to the literature on the differential analysis of Bartonella gltA gene [25,27–29] and Wolbachia 16S rRNA gene in different host [30–33], it was confirmed that Bartonella detected in this study was B. melophagi, 3 of the Wolbachia were supergroup F. Previous reports showed that Bartonella and Wolbachia were detected in sheep keds in Gansu, China [10,11], but Bartonella and Wolbachia were not classified. The sequences of B. melophagi gltA gene (KY224150 and KY224151) and Wolbachia 16S rRNA gene (KY224163 and KY224164) from Xinjiang sheep keds were found in the GenBank database. However, it is short of details in the literature. This is the first study reported that Wolbachia supergroup F found in sheep keds and provided the molecular evidence that sheep keds carried B. melophagi and Wolbachia supergroup F in the Aksu Prefecture of southern Xinjiang, China. The 2 pathogens were found in sheep keds around Taklimakan Desert for the first time.
It is reported previously that Bartonella was detected in sheep and sheep keds [10,11,15,19,20,34]. This study also confirmed the widespread prevalence of B. melophagi in sheep keds in Kuqa County, Aksu Prefecture, China. Currently, no report has revealed the relation between B. melophagi and sheep, but it has been reported that B. bovis was associated with bovine endocarditis [35] and previous reports mentioned that B. melophagi was isolated from 2 female patients with pericarditis and skin lesions with animal exposure history [36]. This study is in agreement with the literature which showed it may be a symbiotic relationship between B. melophagi and sheep keds [19] or B. melophagi is well adapted to sheep and sheep keds, and frequently transferred between 2 hosts [20]. Sheep keds may play an important role in the spread of B. melophagi among sheep. The exact role and mode of action need further study. Close attention should be given that B. melophagi existed in the local population of Xinjiang.
This is the second report in the world that Wolbachia is detected in sheep keds. It also provides another evidence that Wolbachia is the most widely distributed intracellular symbiotic bacteria. Three Wolbachia strains in this study are attributed to supergroup F. It is the first report of the supergroup F Wolbachia on sheep keds. In this study, we need to further analyze and identify the subgroups of 1 strain in combination with other Wolbachia gene fragments. In addition, genetic analysis is based on the Wolbachia 16S rRNA gene. The classification of supergroups B, N, K, and I is relatively confusing, thus, more accurate and detailed classification requires multi-gene tandem or whole genome integrated data analysis. Wolbachia plays a regulatory role in host reproductive behavior by means such as inducing host cytoplasm incompatibility and parthenogenesis [11]. Further studies are needed to determine its role in regulating the reproductive behavior and regulation mode.
Bartonella and Wolbachia are zoonosis pathogens. They use vampire arthropods as their parasitic host, storing hosts or the media. They are highly valued by veterinary medicine and by the medicine in the control of infectious diseases or vector biological control.
ACKNOWLEDGMENTSThis study was funded by the National Natural Science Foundation of China (No. 31460655) and the open project of Key Laboratory of Tarim Animal Husbandry Science and Technology, Xinjiang Production & Construction Corps (HS201501, HS201801).
REFERENCES2. Liu D, Wang YZ, Zhang H, Liu ZQ, Wureli HZ, Wang SW, Tu CC, Chen CF. First report of Rickettsia raoultii and R. slovaca in Melophagus ovinus, the sheep ked. Parasit Vectors 2016;9:600.
3. Bequaert JC. A monograph of the Melophaginae, or ked-flies, of sheep, goats, deer and antelopes (Diptera: Hippoboscidae). Entomol Am 1942;22:1-220.
4. Tetley JH. The sheep ked, Melophagus ovinus L. I. Dissemination potential. Parasitology 1958;48:353-363.
5. Wu HS, Yang N, Ma SM, Chen HJ, Zhen Y, Ge RL. A survey of three zoonosis and ectoparasite of Tibet an antelope. Chin Qinghai J Anim Vet Sci 2010;40:21-22.
6. Izdebska JN. European bison arthropod parasites from closed Polish breeding facilities. Acta Parasitol 2001;46:135-137.
7. Lassnig H, Prosl H, Hinterdorfer F. Parasites of the red fox (Vulpes vulpes) in Styria. Wien Tierarztl Monat 1998;85:116-122.
8. Chu CY, Jiang BG, Qiu EC, Zhang F, Zuo SQ, Yang H, Liu W, Cao WC.
Borrelia burgdorferi sensu lato in sheep keds (Melophagus ovinus), Tibet, China. Vet Microbiol 2011;149:526-529.
9. Jiang WC, Zhang SF, Jin ZY. Diagnosis and treatment of sheep keds (Melophagus ovinus) in sheep. Agric Sci J Yanbian Univ 1983;14:62-64.
10. Duan DY, Liu GH, Cheng TY, Wang YQ. Microbial population analysis of the midgut of Melophagus ovinus via high-throughput sequencing. Parasit Vectors 2017;10:382.
11. Wang YQ, Cheng TY, Duan DY. Identification of the microflora in the midgut and pupa of Melophagus ovinus
. Chin Vet Sci 2017;47:861-865.
12. Wang Y, Sun BJ, Yu CY, Ni X.
Melophagus ovinus Linnaeus intercepted and captured from Canadian pickled goatskins at Linyi port. Chin J Vector Biol Control 2010;21:35.
13. Zhang YZ, Si XL, Gao GB. Anthelmintic effect of sheep keds, woolen lice and imported insect repellents in imported sheep. Chin J Vet Med 1996;22:48-51.
14. Wang W. There are 4 eggs in the imported wool, Shaoxing intercepted exotic pest parasites. [Internet]. Shaoxing evening; 2017. Feb. 12. [cited 2017 Feb 12]. Available from: http://zj.zjol.com.cn/news/554539
15. Kumsa B, Parola P, Raoult D, Socolovschi C.
Bartonella melophagi in Melophagus ovinus (sheep ked) collected from sheep in northern Oromia, Ethiopia. Comp Immunol Microbiol Infect Dis 2014;37:69-76.
16. Gibson W, Pilkington JG, Pemberton JM.
Trypanosoma melophagium from the sheep ked Melophagus ovinus on the island of St Kilda. Parasitology 2010;137:1799-1804.
17. Hornok S, de la Fuente J, Biró N, Fernández de Mera IG, Meli ML, Elek V, Gönczi E, Meili T, Tánczos B, Farkas R, Lutz H, Hofmann-Lehmann R. First molecular evidence of Anaplasma ovis and Rickettsia spp. in keds (Diptera: Hippoboscidae) of sheep and wild ruminants. Vector Borne Zoonotic Dis 2011;11:1319-1321.
18. Luedke AJ, Jochim MM, Bowne JG. Preliminary bluetongue transmission with the sheep ked Melophagus ovinus (L.). Can J Comp Med Vet Sci 1965;29:229-231.
19. Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, Vayssier-Taussat M, Boulouis HJ. Role of Hippoboscidae flies as potential vectors of Bartonella spp. infecting wild and domestic ruminants. Appl Environ Microbiol 2004;70:6302-6305.
20. Kosoy M, Bai Y, Enscore R, Rizzo MR, Bender S, Popov V, Albayrak L, Fofanov Y, Chomel B.
Bartonella melophagi in blood of domestic sheep (Ovis aries) and sheep keds (Melophagus ovinus) from the southwestern US: Cultures, genetic characterization, and ecological connections. Vet Microbiol 2016;190:43-49.
21. Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?--A statistical analysis of current data. FEMS Microbiol Lett 2008;281:215-220.
22. Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 2012;7:e38544.
23. Glowska E, Dragun-Damian A, Dabert M, Gerth M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect Genet Evol 2015;30:140-146.
24. Li DM, Liu QY, Yu DZ, Zhang JZ, Gong ZD, Song XP. Phylogenetic analysis of Bartonella detected in rodent fleas in Yunnan, China. J Wildl Dis 2007;43:609-617.
25. Li DM, Hou Y, Song XP, Fu YQ, Li GC, Li M, Eremeeva ME, Wu HX, Pang B, Yue YJ, Huang Y, Lu L, Wang J, Liu QY. High prevalence and genetic heterogeneity of rodent-borne Bartonella species on Heixiazi Island, China. Appl Environ Microbiol 2015;81:7981-7992.
26. Palomar AM, García-Álvarez L, Santibáñez S, Portillo A, Oteo JA. Detection of tick-borne ‘Candidatus Neoehrlichia mikurensis’ and Anaplasma phagocytophilum in Spain in 2013. Parasit Vectors 2014;7:57.
27. Gil H, García-Esteban C, Barandika JF, Peig J, Toledo A, Escudero R, Jado I, Rodríguez-Vargas M, García-Amil C, Lobo B, Roales P, Rodríguez-Moreno I, Olmeda AS, García-Pérez AL, Anda P. Variability of Bartonella genotypes among small mammals in Spain. Appl Environ Microbiol 2010;76:8062-8070.
28. Rao HX, Yu J, Guo P, Ma YC, Liu QY, Jiao M, Ma ZW, Ge H, Wang CX, Song XP, Shi Y, Li DM.
Bartonella Species Detected in the plateau pikas (Ochotona curzoiae) from Qinghai Plateau in China. Biomed Environ Sci 2015;28:674-678.
29. Mullins KE, Hang J, Clifford RJ, Onmus-Leone F, Yang Y, Jiang J, Leguia M, Kasper MR, Maguina C, Lesho EP, Jarman RG, Richards A, Blazes D. Whole-Genome Analysis of Bartonella ancashensis, a Novel Pathogen Causing verruga peruana, Rural Ancash Region, Peru. Emerg Infect Dis 2017;23:430-438.
30. Dunn AK, Stabb EV. Culture-independent characterization of the microbiota of the ant lion Myrmeleon mobilis (Neuroptera: Myrmeleontidae). Appl Environ Microbiol 2005;71:8784-8794.
31. Augustinos AA, Santos-Garcia D, Dionyssopoulou E, Moreira M, Papapanagiotou A, Scarvelakis M, Doudoumis V, Ramos S, Aguiar AF, Borges PA, Khadem M, Latorre A, Tsiamis G, Bourtzis K. Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PLoS One 2011;6:e28695.
32. Sakamoto JM, Feinstein J, Rasgon JL.
Wolbachia infections in the Cimicidae: museum specimens as an untapped resource for endosymbiont surveys. Appl Environ Microbiol 2006;72:3161-3167.
33. Roy V, Harry M. Diversity of Wolbachia isolated from the Cubitermes sp. affinis subarquatus complex of species (Termitidae), revealed by multigene phylogenies. FEMS Microbiol Lett 2007;274:102-111.
34. Rudolf I, Betášová L, Bischof V, Venclíková K, Blažejová H, Mendel J, Hubálek Z, Kosoy M. Molecular survey of arthropod-borne pathogens in sheep keds (Melophagus ovinus), Central Europe. Parasitol Res 2016;115:3679-3682.
Fig. 1Phylogenetic tree of Bartonella based on gltA gene. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model. The analysis involved 42 nucleotide sequences. There were a total of 309 positions in the final dataset. Evolutionary analyses were conducted in MEGA5. Sequences of this work were marked with black triangle (▲). 
Fig. 2Phylogenetic tree of Wolbachia based on 16S rRNA gene. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model. The analysis involved 40 nucleotide sequences. There were a total of 239 positions in the final dataset. Evolutionary analyses were conducted in MEGA5. Sequences of this work were marked with black circular (●). 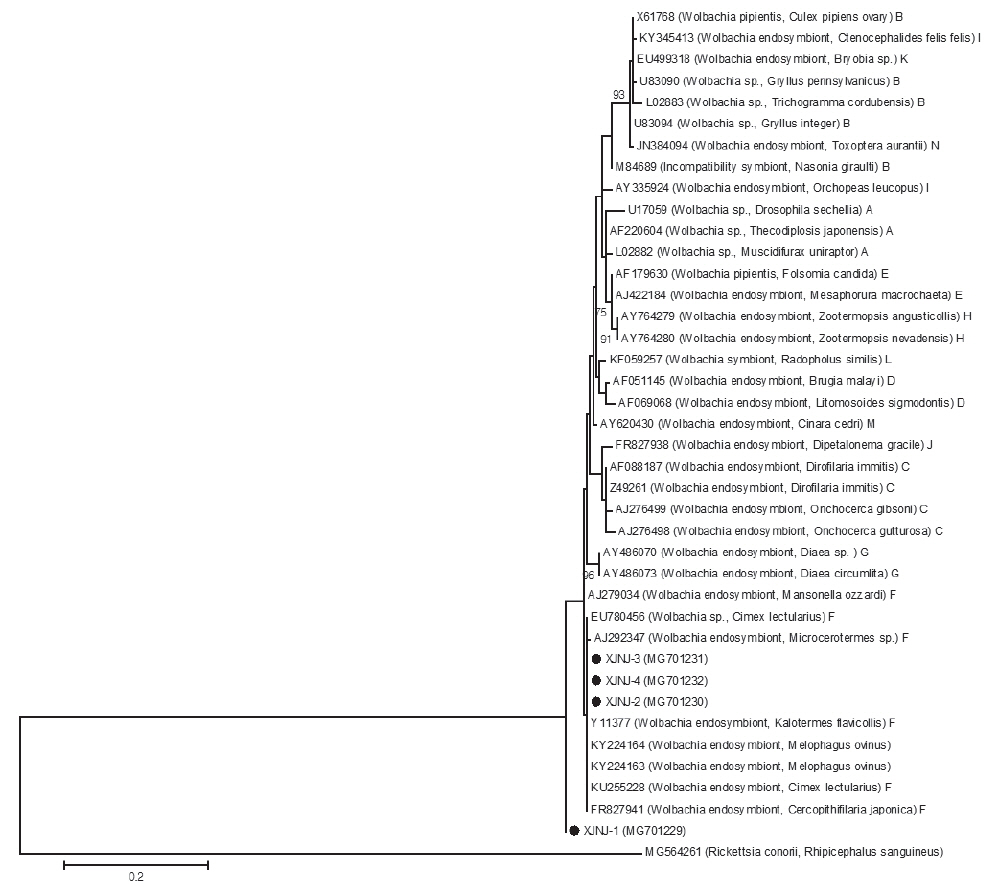
Table 1BLAST comparison analysis results of Bartonella gltA gene and Wolbachia 16S rRNA gene |
|
|||||||||||||||||||||||||||||||||||||||||